Research
In recent years, genomic sequencing and systematic analysis have revealed many molecular components that control gene expression at multiple levels, and detailed the myriad of interactions among and between these regulatory modules. However, it remains unclear how these molecular networks operate in time and space to carry out diverse cellular functions. The goal of our lab is to understand how network architecture governs the dynamics and function of regulatory responses in the context of stresses, aging, or diseases.
Our lab uses multidisciplinary approaches, integrating biology, engineering and computer science, in our studies. We develop and use novel microfluidics and imaging technologies to quantitatively track the dynamics of molecular processes in single living cells and construct computational models to describe and predict cellular behaviors.
In particular, our laboratory focuses on the following three major research themes:
Decoding the dynamics of stress responses
Cells could transmit environmental information and initiate cellular responses through regulating the temporal dynamics of signaling activities. In an oft-cited example, epidermal growth factor induces transient activation of the mitogen activated protein kinase ERK and leads to cell proliferation, whereas nerve growth factor elicits sustained ERK activation and results in cell differentiation. In S.cerevisiae, we have recently discovered that the general stress responsive transcription factor Msn2 encodes both the identity and strength of external stimuli into dynamic patterns of its nuclear translocation. We further revealed that these dynamic patterns could lead to selective gene expression. Our current work focuses on understanding: (1) the network structures that encode environmental information into signaling dynamics; (2) the gene circuits
that decode signaling dynamics into functions; (3) the mechanisms that underlie cellular memory of environmental changes.
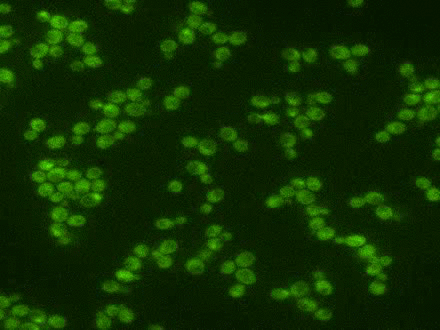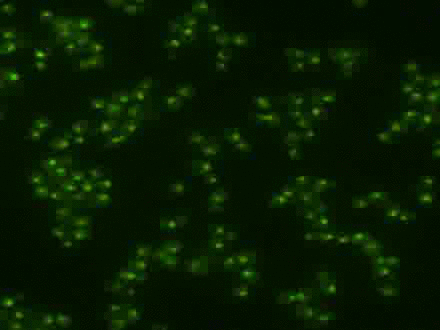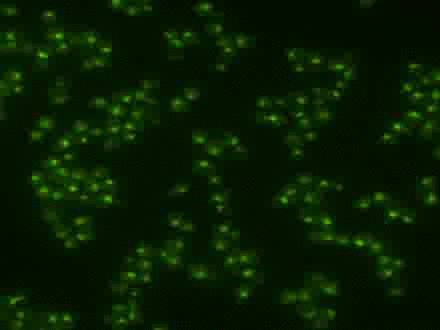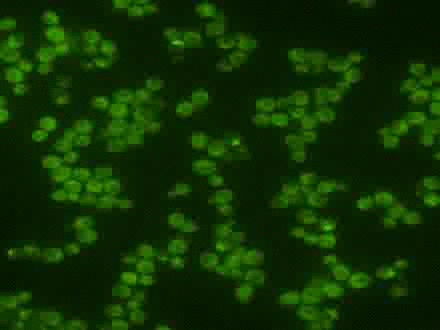
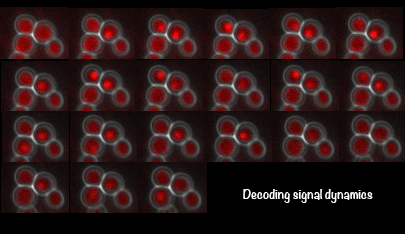
Using the signal control system to generate oscillatory transcription factor activation
A synthetic yeast strain has been constructed by introducing analog-sensitive mutations in PKA that renders selective inhibition of PKA activity by a small cell-permeable inhibitor, 1-NM-PP1. This chemical genetic strategy was combined with microfluidics and quantitative microscopy to control dynamic patterns of PKA input and monitor single-cell responses. This system was used to systematically investigate how the dynamics of transcription factor Msn2 are interpreted by its target genes (Hao et al., 2013; Hao and O'Shea, 2012). For example, in the movie, oscillatory patterns of transcription factor translocation were artificially generated using the synthetic system.
Probing the causes of cellular aging
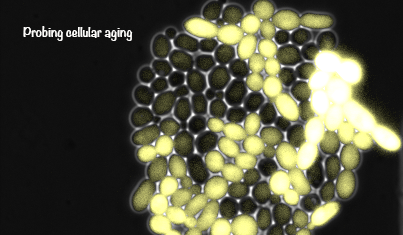
Cellular aging plays an important role in many diseases, such as cancers and neurodegenerative disorders. Although there has been steady progress in identifying age-related factors such as reactive oxygen species and genomic instability, an emerging challenge is to reconcile the contributions of these factors with the fact that genetically identical cells age at significantly different rates. Such complexity requires single-cell analyses designed to unravel the interplay of aging dynamics and variability. We developed novel microfluidic technologies that enable tracking of the replicative aging of single yeast cells in a fully automated fashion. We aim to combine this transformative approach with computational modeling to obtain a quantitative and predictive understanding of the aging process and to develop new strategies to promote longevity.
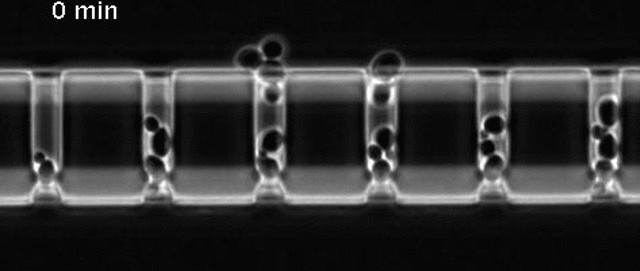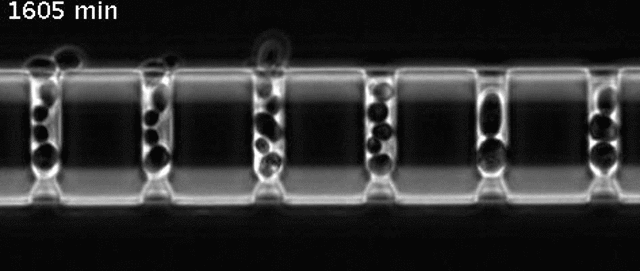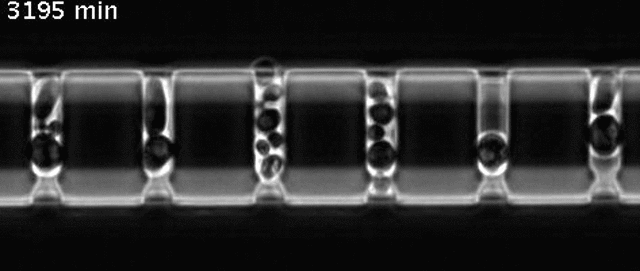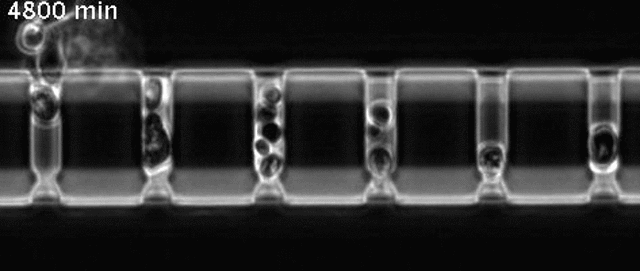
Yeast cells aging in a microfluidic chip
The aging mother cells are trapped at the bottom of finger-shaped chambers, producing daughter cells until their final death.

Divergent aging trajectories of isogenic cells
Ribosomal DNA-GFP reporter and nuc. iRFP (heme) reporter were co-expressed in the same cells and their fluorescence were measured during aging. After the aging trajectories of the two representative cells were quantified and plotted, trajectories of a population of isogenic WT cells were plotted in the space (Mode 1 – red; Mode 2 – blue). See Li et al. Science 2020 for details.
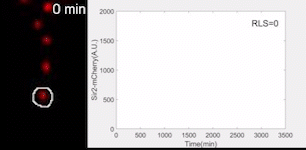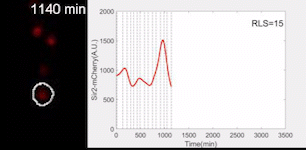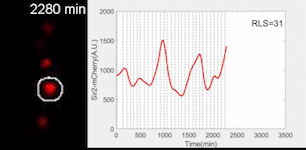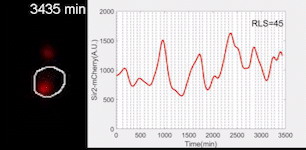
Sir2 abundance in the synthetic gene oscillator during aging
The movie of a representative engineered cell in which Sir2-mCherry fluorescence were measured across the lifespan and the real-time quantification of reporter fluorescence was plotted as a function of time. See Zhou et al. Science 2023 for details.
Quantifying the heterogeneity in cancer cells
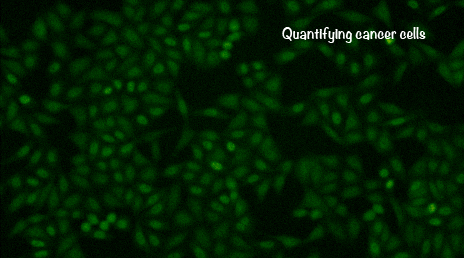
We have expanded our quantitative biology research into mammalian cell systems. We have developed new microfluidic and imaging technologies to track individual mammalian cells over a very long period of time. Our current work focuses on studying the heterogeneous signaling dynamics in cancer cells and how these dynamics influence cancer cell growth.
Tracing the interferon responses in single HeLa cells
STAT1 nuclear translocation and IRF9 expression were monitored in the same cells in response to 100 ng/mL interferon-alpha. Cell-to-cell variation has been observed. See Mudla et al. eLife 2020 for details.